Use case: Building a medical intelligence application with augmented patient data
Generative AI can help augment patient care and staff productivity by enhancing both clinical and administrative functions. AI-driven image analysis, such as interpreting sonograms, accelerates diagnostic processes and improves accuracy. It can provide critical insights that support timely medical interventions.
When you combine generative AI models with knowledge graphs, you can automate the chronological organization of electronic patient records. This helps you integrate real-time data from doctor-patient interactions, symptoms, diagnoses, lab results, and image analysis. This equips the doctor with comprehensive patient data. This data helps the doctor make more accurate and timely medical decisions, enhancing both patient outcomes and healthcare provider productivity.
Solution overview
AI can empower doctors and clinicians by synthesizing patient data and medical knowledge to provide valuable insights. This Retrieval Augmented Generation (RAG) solution is a medical intelligence engine that consumes a comprehensive set of patient data and knowledge from millions of clinical interactions. It harnesses the power of generative AI to create evidence-based insights for improved patient care. It is designed to enhance the clinical workflows, reduce errors, and improve patient outcomes.
The solution includes an automated image-processing capability that is powered by LLMs. This capability reduces the amount of time that medical personnel must spend manually searching for similar diagnostic images and analyzing diagnostic results.
The following image shows the end-to-end-workflow for this solution. It uses Amazon Neptune, Amazon SageMaker AI, Amazon OpenSearch Service, and a foundation model in Amazon Bedrock. For the context retrieval agent that interacts with the medical knowledge graph in Neptune, you can choose between an Amazon Bedrock agent and a LangChain agent.

In our experiments with sample medical questions, we observed that the final responses generated by our approach using knowledge graph maintained in Neptune, OpenSearch vector database housing clinical knowledge base, and Amazon Bedrock LLMs were grounded in factuality and are far more accurate by reducing the false positives and boosting the true positives. This solution can generate evidence-based insights on patient's health status and aims to enhance the clinical workflows, reduce errors, and improve patient outcomes.
Building this solution consists of the following steps:
Step 1: Discovering data
There are many open source medical datasets that you can use to support the
development of a healthcare AI-driven solution. One such dataset is the MIMIC-IV dataset
You might also use a dataset that provides annotated, de-identified patient discharge summaries that are specifically curated for research purposes. A discharge summary dataset can help you experiment with entity extraction, allowing you to identify key medical entities (such as conditions, procedures, and medications) from the text. Step 2: Building a medical knowledge graph in this guide describes how you can use the structured data extracted from the MIMIC-IV and discharge summary datasets to create a medical knowledge graph. This medical knowledge graph serves as the backbone for advanced querying and decision-support systems for healthcare professionals.
In addition to text-based datasets, you can use image datasets. For example, the
Musculoskeletal
Radiographs (MURA) dataset
Step 2: Building a medical knowledge graph
For any healthcare organization that wants to build a decision-support system based on a massive knowledge base, a key challenge is to locate and extract the medical entities that are present in the clinical notes, medical journals, discharge summaries, and other data sources. You also need to capture the temporal relationships, subjects, and certainty assessments from these medical records in order to effectively use the extracted entities, attributes, and relationships.
The first step is to extract medical concepts from the unstructured medical text by using a few-shot prompt for a foundation model, such as Llama 3 in Amazon Bedrock. Few-shot prompting is when you provide an LLM with a small number of examples that demonstrate the task and desired output before asking it to perform a similar task. Using an LLM-based medical entity extractor, you can parse the unstructured medical text and then generate a structured data representation of the medical knowledge entities. You can also store the patient attributes for downstream analysis and automation. The entity extraction process includes the following actions:
-
Extract information about medical concepts, such as diseases, medications, medical devices, dosage, medicine frequency, medicine duration, symptoms, medical procedures, and their clinically relevant attributes.
-
Capture functional features, such as temporal relationships between extracted entities, subjects, and certainty assessments.
-
Expand standard medical vocabularies, such as the following:
-
Concept identifiers (RxCUI) from the RxNorm database
-
Codes from the International Classification of Diseases, 10th Revision, Clinical Modification (ICD-10-CM)
-
Terms from Medical Subject Headings (MeSH)
-
Concepts from Systematized Nomenclature of Medicine, Clinical Terms (SNOMED CT)
-
Codes from Unified Medical Language System (UMLS)
-
-
Summarize discharge notes and derive medical insights from transcripts.
The following figure shows the entity extraction and schema validation steps to create valid paired combinations of entities, attributes, and relationships. You can store unstructured data, such as discharge summaries or patient notes, in Amazon Simple Storage Service (Amazon S3). You can store structured data, such as enterprise resource planning (ERP) data, electronic patient records, and lab information systems, in Amazon Redshift and Amazon DynamoDB. You can build an Amazon Bedrock entity-creation agent. This agent can integrate services, such as Amazon SageMaker AI data-extraction pipelines, Amazon Textract, and Amazon Comprehend Medical, to extract entities, relationships, and attributes from the structured and unstructured data sources. Finally, you use an Amazon Bedrock schema-validation agent to make sure that the extracted entities and relationships conform to the predefined graph schema and maintain the integrity of node-edge connections and associated properties.
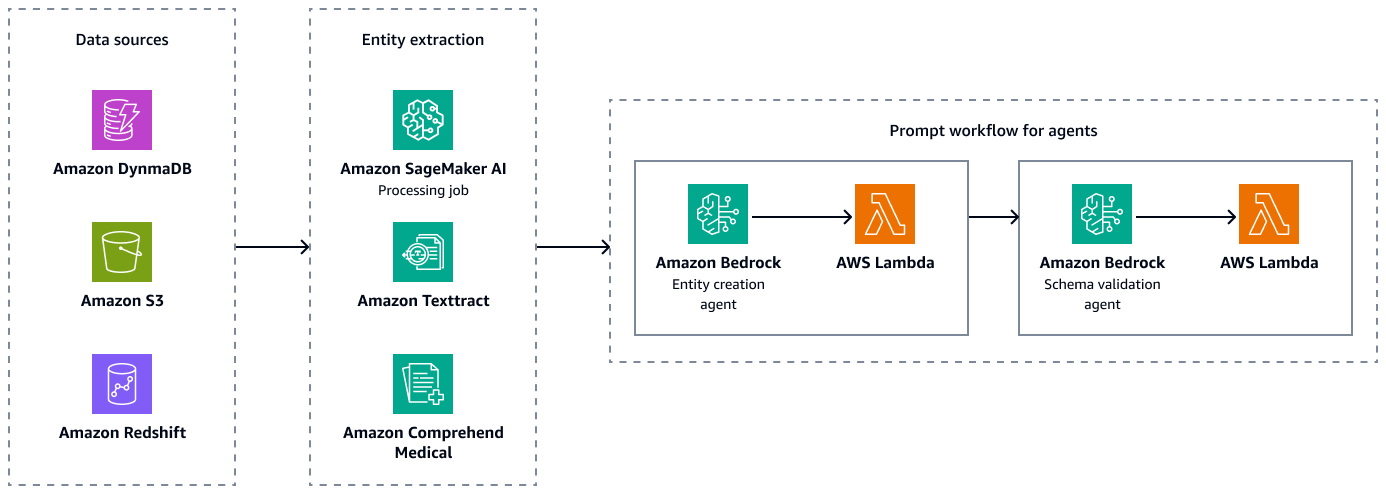
After extraction and validation of the entities, relations, and attributes, you can link them to create a subject-object-predicate triplet. You ingest this data into an Amazon Neptune graph database, as shown in the following figure. Graph databases are optimized to store and query the relationships between data items.
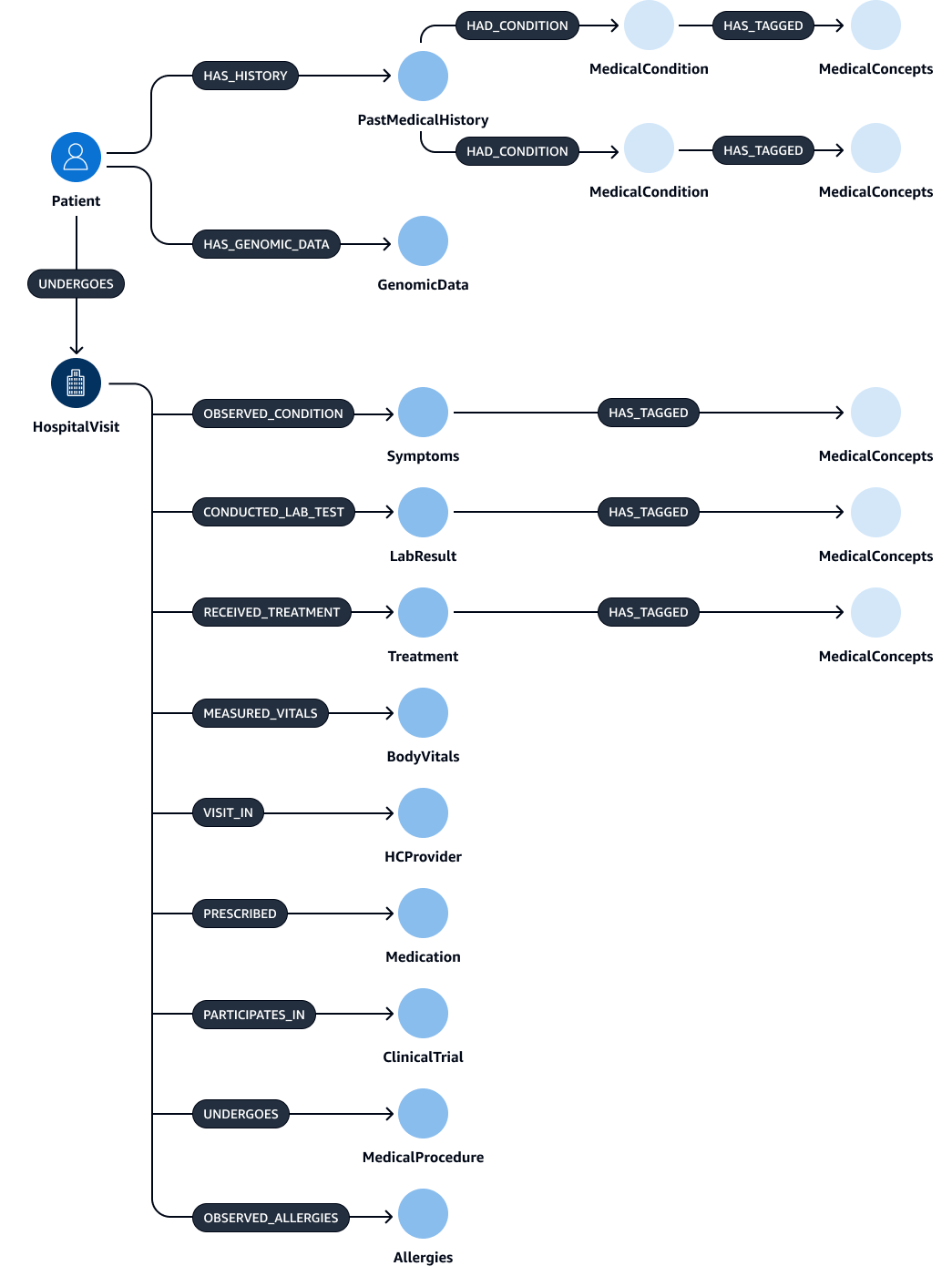
You could create a comprehensive knowledge graph with this data. A knowledge graphHospitalVisit,
PastMedicalHistory, Symptoms, Medication,
MedicalProcedures, and Treatment.
The following tables lists the entities and their attributes that you might extract from discharge notes.
| Entity | Attributes |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
The following table lists the relationships that entities might have and their
corresponding attributes. For example, the Patient entity might connect to
the HospitalVisit entity with the [UNDERGOES] relationship.
The attribute for this relationship is VisitDate.
| Subject entity | Relationship | Object entity | Attributes |
|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
None |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
None |
|
|
|
None |
|
|
|
None |
|
|
|
None |
Step 3: Building context retrieval agents to query the medical knowledge graph
After you build the medical graph database, the next step is to build agents for graph interaction. The agents retrieve the correct and required context for the query that a doctor or clinician inputs. There are several options for configuring these agents that retrieve the context from the knowledge graph:
Amazon Bedrock agents for graph interaction
Amazon Bedrock agents work seamlessly with Amazon Neptune graph databases. You can perform advanced interactions through Amazon Bedrock action groups. The action group initiates the process by calling an AWS Lambda function, which runs Neptune openCypher queries.
For querying a knowledge graph, you can use two distinct approaches: direct query execution or querying with context embedding. These approaches can be applied independently or combined, depending on your specific use case and ranking criteria. By combining both approaches, you can provide more comprehensive context to the LLM, which can improve results. The following are the two query execution approaches:
-
Direct Cypher query execution without embeddings – The Lambda function executes queries directly against Neptune without any embeddings-based search. The following is an example of this approach:
MATCH (p:Patient)-[u:UNDERGOES]->(h:HospitalVisit) WHERE h.Reason = 'Acute Diabetes' AND date(u.VisitDate) > date('2024-01-01') RETURN p.PatientID, p.Name, p.Age, p.Gender, p.Address, p.ContactInformation -
Direct Cypher query execution using embedding search – The Lambda function uses embedding search to enhance the query results. This approach enhances the query execution by incorporating embeddings, which are dense vector representations of data. Embeddings are particularly useful when the query requires semantic similarity or broader understanding beyond exact matches. You can use pre-trained or custom-trained models to generate embeddings for each medical condition. The following is an example of this approach:
CALL { WITH "Acute Diabetes" AS query_term RETURN search_embedding(query_term) AS similar_reasons } MATCH (p:Patient)-[u:UNDERGOES]->(h:HospitalVisit) WHERE h.Reason IN similar reasons AND date(u.VisitDate) > date('2024-01-01') RETURN p.PatientID, p.Name, p.Age, p.Gender, p.Address, p.ContactInformationIn this example, the
search_embedding("Acute Diabetes")function retrieves conditions that are semantically close to "Acute Diabetes." This helps the query to also find patients that have conditions such as pre-diabetes or metabolic syndrome.
The following image shows how Amazon Bedrock agents interact with Amazon Neptune in order to perform a Cypher query of a medical knowledge graph.

The diagram shows the following workflow:
-
The user submits a question to the Amazon Bedrock agent.
-
The Amazon Bedrock agent passes the question and input filter variables to the Amazon Bedrock action groups. These action groups contain an AWS Lambda function that interacts with the Amazon SageMaker AI text embedding endpoint and the Amazon Neptune medical knowledge graph.
-
The Lambda function integrates with the SageMaker AI text embedding endpoint to perform a semantic search within the openCypher query. It converts the natural language query into an openCypher query by using underlying LangChain agents.
-
The Lambda function queries the Neptune medical knowledge graph for the correct dataset and receives the output from the Neptune medical knowledge graph.
-
The Lambda function returns the results from Neptune to the Amazon Bedrock action groups.
-
The Amazon Bedrock action groups send the retrieved context to the Amazon Bedrock agent.
-
The Amazon Bedrock agent generates the response by using the original user query and the retrieved context from the knowledge graph.
LangChain agents for graph interaction
You can integrate LangChain with Neptune to enable graph-based queries and retrievals. This approach can enhance AI-driven workflows by using the graph database capabilities in Neptune. The custom LangChain retriever acts as an intermediary. The foundational model in Amazon Bedrock can interact with Neptune by using both direct Cypher queries and more complex graph algorithms.
You can use the custom retriever to refine how the LangChain agent interacts with the Neptune graph algorithms. For example, you can use few-shot prompting, which helps you tailor the foundation model's responses based on specific patterns or examples. You can also apply LLM-identified filters to refine the context and improve the precision of responses. This can improve the efficiency and accuracy of the overall retrieval process when interacting with complex graph data.
The following image shows how a custom LangChain agent orchestrates the interaction between an Amazon Bedrock foundation model and an Amazon Neptune medical knowledge graph.

The diagram shows the following workflow:
-
A user submits a question to Amazon Bedrock and the LangChain agent.
-
The Amazon Bedrock foundation model uses the Neptune schema, which is provided by the LangChain agent, to generate a query for the user's question.
-
The LangChain agent runs the query against the Amazon Neptune medical knowledge graph.
-
The LangChain agent sends the retrieved context to the Amazon Bedrock foundation model.
-
The Amazon Bedrock foundation model uses the retrieved context to generate an answer to the user's question.
Step 4: Creating a knowledge base of real-time, descriptive data
Next, you create a knowledge base of real-time, descriptive doctor-patient interaction
notes, diagnostic image assessments, and lab analysis reports. This knowledge base is a
vector database
Using an OpenSearch Service medical knowledge base
Amazon OpenSearch Service can manage large volumes of high-dimensional medical data. It is a managed service that facilitates high-performance search and real-time analytics. It is well suited as a vector database for RAG applications. OpenSearch Service acts as a backend tool to manage vast amounts of unstructured or semi-structured data, such as medical records, research articles, and clinical notes. Its advanced semantic search capabilities help you retrieve contextually relevant information. This makes it particularly useful in applications such as clinical decision-support systems, patient query resolution tools, and healthcare knowledge management systems. For instance, a clinician can quickly find relevant patient data or research studies that match specific symptoms or treatment protocols. This helps clinicians make decisions that are informed by the most up-to-date and relevant information.
OpenSearch Service can scale and handle real-time data indexing and querying. This makes it ideal for dynamic healthcare environments where timely access to accurate information is critical. Additionally, it has multi-modal search capabilities that are optimal for searches that require multiple inputs, such as medical images and doctor notes. When implementing OpenSearch Service for healthcare applications, it is crucial that you define precise fields and mappings in order to optimize data indexing and retrieval. Fields represent the individual pieces of data, such as patient records, medical histories, and diagnostic codes. Mappings define how these fields are stored (in embedding form or original form) and queried. For healthcare applications, it is essential to establish mappings that accommodate various data types, including structured data (such as numerical test results), semi-structured data (such as patient notes), and unstructured data (such as medical images)
In OpenSearch Service, you can perform full-text neural
search
Creating a RAG architecture
You can deploy a customized RAG solution that uses Amazon Bedrock agents to query a medical knowledge base in OpenSearch Service. To accomplish this, you create an AWS Lambda function that can interact with and query OpenSearch Service. The Lambda function embeds the user's input question by accessing a SageMaker AI text embedding endpoint. The Amazon Bedrock agent passes additional query parameters as inputs to the Lambda function. The function queries the medical knowledge base in OpenSearch Service, which returns the relevant medical content. After you set up the Lambda function, add it as an action group within the Amazon Bedrock agent. The Amazon Bedrock agent takes the user's input, identifies the necessary variables, passes the variables and question to the Lambda function, and then initiates the function. The function returns a context that helps the foundation model provide a more accurate answer to the user's question.

The diagram shows the following workflow:
-
A user submits a question to the Amazon Bedrock agent.
-
The Amazon Bedrock agent selects which action group to initiate.
-
The Amazon Bedrock agent initiates an AWS Lambda function and passes parameters to it.
-
The Lambda function initiates the Amazon SageMaker AI text embedding model to embed the user question.
-
The Lambda function passes the embedded text and additional parameters and filters to Amazon OpenSearch Service. Amazon OpenSearch Service queries the medical knowledge base and returns results to the Lambda function.
-
The Lambda function passes the results back to the Amazon Bedrock agent.
-
The foundation model in the Amazon Bedrock agent generates a response based on the results and returns the response to the user.
For situations where more complex filtering is involved, you can use a custom LangChain retriever. Create this retriever by setting up an OpenSearch Service vector search client that is loaded directly into LangChain. This architecture allows you to pass more variables in order to create the filter parameters. After the retriever is set up, use the Amazon Bedrock model and retriever to set up a retrieval question-answering chain. This chain orchestrates the interaction between the model and retriever by passing the user input and potential filters to the retriever. The retriever returns relevant context that helps the foundation model answer the user's question.
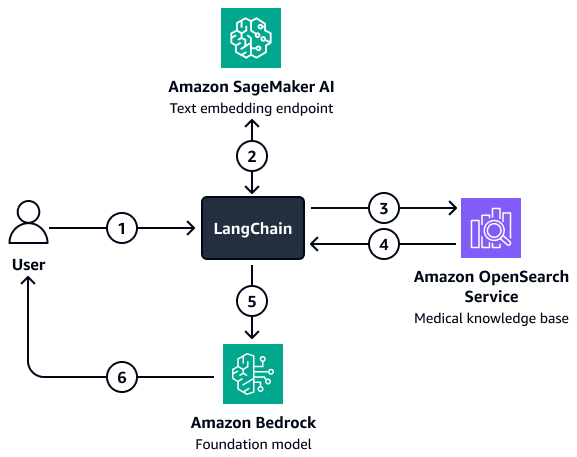
The diagram shows the following workflow:
-
A user submits a question to the LangChain retriever agent.
-
The LangChain retriever agent sends the question to the Amazon SageMaker AI text embedding endpoint to embed the question.
-
The LangChain retriever agent passes the embedded text to Amazon OpenSearch Service.
-
Amazon OpenSearch Service returns the retrieved documents to the LangChain retriever agent.
-
The LangChain retriever agent passes the user question and retrieved context to the Amazon Bedrock foundation model.
-
The foundation model generates a response and sends it to the user.
Step 5: Using LLMs to answer medical questions
The previous steps help you build a medical intelligence application that can fetch a patient's medical records and summarize relevant medications and potential diagnoses. Now, you build the generation layer. This layer uses the generative capabilities of an LLM in Amazon Bedrock, such as Llama 3, to augment the application's output.
When a clinician inputs a query, the context retrieval layer of the application performs the retrieval process from the knowledge graph and returns the top records that pertain to the patient's history, demographics, symptoms, diagnosis, and outcomes. From the vector database, it also retrieves real-time, descriptive doctor-patient interaction notes, diagnostic image assessment insights, lab analysis report summaries, and insights from huge corpus of medical research and academic books. These top retrieved results, the clinician's query, and the prompts (which are tailored to curate answers based on the nature of the query), are then passed to the foundation model in Amazon Bedrock. This is the response generation layer. The LLM uses the retrieved context to generate a response to the clinician's query. The following figure shows the end-to-end workflow of the steps in this solution.

You can use a pre-trained foundational model in Amazon Bedrock, such as Llama 3, for a range of use cases that the medical intelligence application has to handle. The most effective LLM for a given task varies depending on the use case. For example, a pre-trained model might be sufficient to summarize patient-doctor conversations, search through medications and patient histories, and retrieve insights from internal medical data sets and bodies of scientific knowledge. However, a fine-tuned LLM might be necessary for other complex use cases, such as real-time laboratory evaluations, medical procedure recommendations, and predictions of patient outcomes. You can fine-tune an LLM by training it on medical domain datasets. Specific or complex healthcare and life sciences requirements drive development of these fine-tuned models.
For more information about fine-tuning an LLM or choosing an existing LLM that has been trained on medical domain data, see Using large language models for healthcare and life science use cases.
Alignment to the AWS Well-Architected Framework
The solution aligns with all six pillars of the AWS Well-Architected Framework
-
Operational excellence – The architecture is decoupled for efficient monitoring and updates. Amazon Bedrock agents and AWS Lambda help you quickly deploy and roll back tools.
-
Security – This solution is designed to comply with healthcare regulations, such as HIPAA. You can also implement encryption, fine-grained access control, and Amazon Bedrock guardrails to help protect patient data.
-
Reliability – AWS managed services, such as Amazon OpenSearch Service and Amazon Bedrock, provide the infrastructure for continuous model interaction.
-
Performance efficiency – The RAG solution retrieves relevant data quickly by using optimized semantic search and Cypher queries, while an agent router identifies optimal routes for user queries.
-
Cost optimization – The pay-per-token model in Amazon Bedrock and RAG architecture reduce inference and pre-training costs.
-
Sustainability – Using serverless infrastructure and pay-per-token compute minimizes resource usage and enhances sustainability.