本文属于机器翻译版本。若本译文内容与英语原文存在差异,则一律以英文原文为准。
优化用户定义的函数
用户定义的函数 (UDFs) 和 RDD.map in PySpark 通常会显著降低性能。这是因为在 Spark 的底层 Scala 实现中准确表示你的 Python 代码需要开销。
下图显示了 PySpark 作业的架构。当你使用时 PySpark,Spark 驱动程序会使用 Py4J 库从 Python 中调用 Java 方法。在调用 Spark SQL 或 DataFrame 内置函数时,Python 和 Scala 之间几乎没有性能差异,因为这些函数使用优化的执行计划在每个执行器的 JVM 上运行。
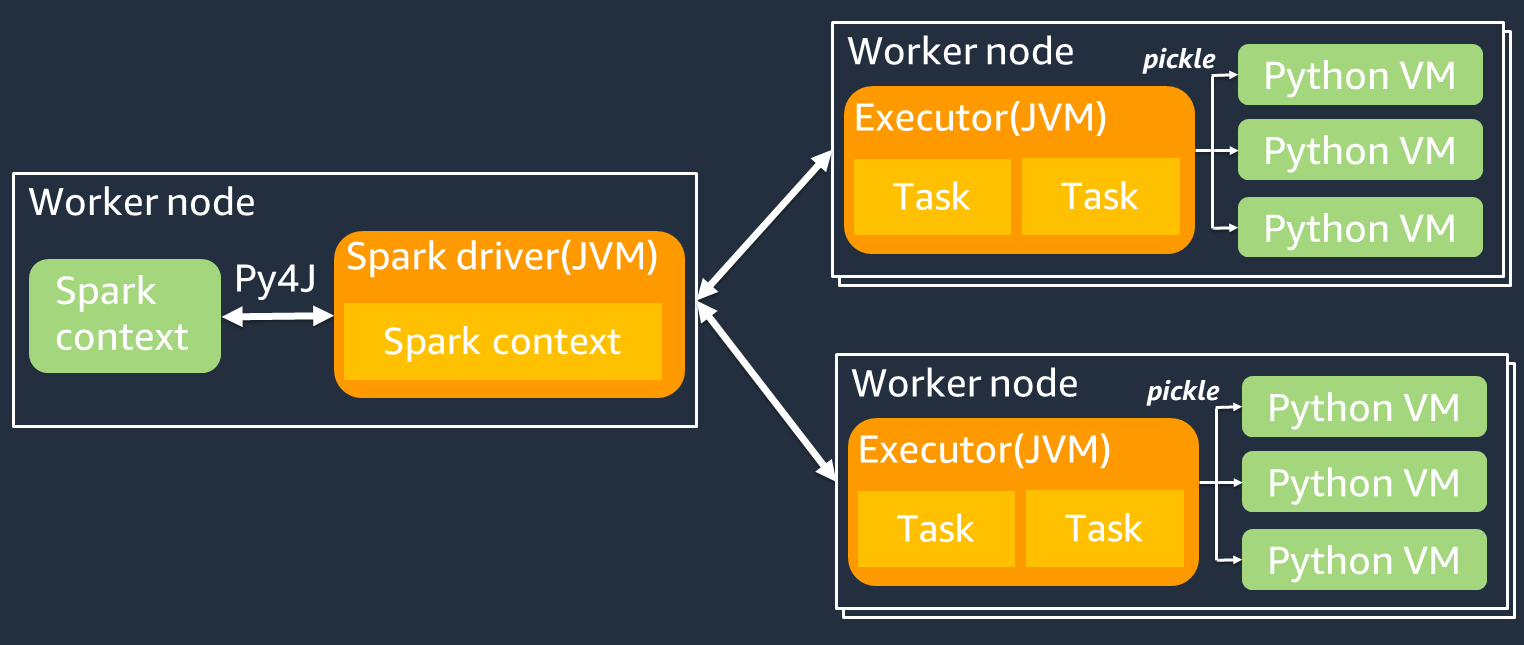
如果您使用自己的 Python 逻辑(例如使用)map/ mapPartitions/ udf,则该任务将在 Python 运行时环境中运行。管理两个环境会产生管理成本。此外,必须对内存中的数据进行转换,以供 JVM 运行时环境的内置函数使用。Pickle 是一种序列化格式,默认用于 JVM 和 Python 运行时之间的交换。但是,这种序列化和反序列化的成本非常高,因此用 Java 或 Scala UDFs 编写的速度比 Python 快。 UDFs
为避免序列化和反序列化开销 PySpark,请考虑以下几点:
-
使用内置的 Spark SQL 函数 — 考虑用 Spark SQL 或 DataFrame内置函数替换你自己的 UDF 或映射函数。在运行 Spark SQL 或 DataFrame 内置函数时,Python 和 Scala 之间的性能差异很小,因为任务是在每个执行器的 JVM 上处理的。
-
使用 Scala 或 Java 实现 UDFs — 考虑使用用 Java 或 Scala 编写的 UDF,因为它们在 JVM 上运行。
-
使用基于 Apache Arrow 的矢量化工作负载 — 考虑使用基 UDFs 于 Arrow 的内容。 UDFs此功能也称为矢量化 UDF (Pandas UDF)。Apache Arrow
是一种与语言无关的内存数据格式, AWS Glue 可用于在 JVM 和 Python 进程之间高效地传输数据。目前,这对使用 Pandas 或 NumPy数据的 Python 用户最有利。 箭头是一种柱状(矢量化)格式。它的使用不是自动的,可能需要对配置或代码进行一些细微的更改才能充分利用并确保兼容性。有关更多详细信息和限制,请参阅中的 Apache Arrow。 PySpark
以下示例比较了标准 Python 中的基本增量 UDF、矢量化 UDF 和 Spark SQL 中的基本增量 UDF。
标准 Python UDF
示例时间为 3.20(秒)。
示例代码
# DataSet df = spark.range(10000000).selectExpr("id AS a","id AS b") # UDF Example def plus(a,b): return a+b spark.udf.register("plus",plus) df.selectExpr("count(plus(a,b))").collect()
执行计划
== Physical Plan == AdaptiveSparkPlan isFinalPlan=false +- HashAggregate(keys=[], functions=[count(pythonUDF0#124)]) +- Exchange SinglePartition, ENSURE_REQUIREMENTS, [id=#580] +- HashAggregate(keys=[], functions=[partial_count(pythonUDF0#124)]) +- Project [pythonUDF0#124] +- BatchEvalPython [plus(a#116L, b#117L)], [pythonUDF0#124] +- Project [id#114L AS a#116L, id#114L AS b#117L] +- Range (0, 10000000, step=1, splits=16)
矢量化的 UDF
示例时间为 0.59(秒)。
矢量化 UDF 的速度比前面的 UDF 示例快 5 倍。你可以看到 Physical PlanArrowEvalPython,这表明这个应用程序是由 Apache Arrow 向量化的。要启用矢量化 UDF,必须在代码spark.sql.execution.arrow.pyspark.enabled = true中指定。
示例代码
# Vectorized UDF from pyspark.sql.types import LongType from pyspark.sql.functions import count, pandas_udf # Enable Apache Arrow Support spark.conf.set("spark.sql.execution.arrow.pyspark.enabled", "true") # DataSet df = spark.range(10000000).selectExpr("id AS a","id AS b") # Annotate pandas_udf to use Vectorized UDF @pandas_udf(LongType()) def pandas_plus(a,b): return a+b spark.udf.register("pandas_plus",pandas_plus) df.selectExpr("count(pandas_plus(a,b))").collect()
执行计划
== Physical Plan == AdaptiveSparkPlan isFinalPlan=false +- HashAggregate(keys=[], functions=[count(pythonUDF0#1082L)], output=[count(pandas_plus(a, b))#1080L]) +- Exchange SinglePartition, ENSURE_REQUIREMENTS, [id=#5985] +- HashAggregate(keys=[], functions=[partial_count(pythonUDF0#1082L)], output=[count#1084L]) +- Project [pythonUDF0#1082L] +- ArrowEvalPython [pandas_plus(a#1074L, b#1075L)], [pythonUDF0#1082L], 200 +- Project [id#1072L AS a#1074L, id#1072L AS b#1075L] +- Range (0, 10000000, step=1, splits=16)
Spark SQL
示例时间为 0.087(秒)。
Spark SQL 比矢量化 UDF 快得多,因为任务是在每个执行者的 JVM 上运行的,没有 Python 运行时。如果你能用内置函数替换 UDF,我们建议你这样做。
示例代码
df.createOrReplaceTempView("test") spark.sql("select count(a+b) from test").collect()
使用熊猫来处理大数据
如果你已经熟悉熊猫